A place for users and developers of the Xataface to discuss and receive support.
 by krazykit » Mon Dec 10, 2012 10:55 am by krazykit » Mon Dec 10, 2012 10:55 am
Software: Xataface 2.0alpha1
Issue: Unable to set "Default Values" during data import on the Import Records Form
I've managed to write an import filter and make Xataface read it in correctly. The imported records will show up in the preview pane just fine, but I can't set any default values when I'm trying to import. The "Default Values" just shows the headers "Key" and "Value", but no way to enter any data (I'd post a screenshot, but phpbb is whining that it looks too spammy).
I don't see any reason that this wouldn't show up: is there something that I'm missing in an ini file or something? The Import Filters how-to page suggests that it should "just work". I can upload ini files or code as needed. Thanks in advance.
-
krazykit
-
- Posts: 4
- Joined: Mon Dec 10, 2012 10:28 am
 by shannah » Mon Dec 10, 2012 11:25 am by shannah » Mon Dec 10, 2012 11:25 am
Can you post your import filter code? Are you specifying the default values at using the import form at the time that you perform the import? Or are you referring to default values that are specified at the database definition level (or the fieldname__default() method of the delegate class)?
-Steve
-
shannah
-
- Posts: 4457
- Joined: Wed Dec 31, 1969 5:00 pm
 by krazykit » Mon Dec 10, 2012 12:01 pm by krazykit » Mon Dec 10, 2012 12:01 pm
- Code: Select all
<?
import('tables/common/fasta.php');
class tables_genesequences {
function __import__fasta(&$data, $defaultValues=array()){
#declare the dataface records
$app =& Dataface_Application::getInstance();
$output = array();
$record = new Dataface_Record('genesequences', array());
$record->setValues($defaultValues); #set the defaults for dataface
$records = array();
#run the data through read_fasta for an array with the data
#array( $header, $sequence);
$output = read_fasta($data);
//well, i know how to do this as a loop...
foreach($output as $vals)
{
$record -> setValues( array(
'header' => $vals['header'],
'sequence' => $vals['sequence']
)
);
$records[] = $record;
}
return $records;
}
}
?>
the read_fasta() function simply returns an array with an array for each header,sequence pair. edit: here's my fasta.php, not that it should matter: - Code: Select all
<?php
/* ex. fasta: multiple sequences are simply appended
>gi|5524211|gb|AAD44166.1| cytochrome b [Elephas maximus maximus]
LCLYTHIGRNIYYGSYLYSETWNTGIMLLLITMATAFMGYVLPWGQMSFWGATVITNLFSAIPYIGTNLV
EWIWGGFSVDKATLNRFFAFHFILPFTMVALAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLG
LLILILLLLLLALLSPDMLGDPDNHMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGVLALFLSIVIL
GLMPFLHTSKHRSMMLRPLSQALFWTLTMDLLTLTWIGSQPVEYPYTIIGQMASILYFSIILAFLPIAGX
IENY
*/
//read & analyse a fasta string - rewriten by pb
function beginsWith( $str, $sub ) {
return ( substr( $str, 0, strlen( $sub ) ) === $sub );
}
function read_fasta( $source )
{
// split each line
$source_array = explode("\n", $source);
$pos = -1;
$seqinfo = array();
foreach ($source_array as $line)
{
// do we find > .... then this is a new sequence
$new_sequence = strpos($line, ">");
$new_note = strpos($line, ";"); # older
if( beginsWith($line, ">") || beginsWith($line, ";"))
{
$pos = $pos +1;
$seqinfo[] = array('header' => $line, 'sequence' => '');
}
else
{
$seqinfo[$pos]["sequence"] .= $line;
}
}
return $seqinfo;
}
?>
-
krazykit
-
- Posts: 4
- Joined: Mon Dec 10, 2012 10:28 am
 by shannah » Mon Dec 10, 2012 4:06 pm by shannah » Mon Dec 10, 2012 4:06 pm
Initialize the record inside the array: - Code: Select all
$record = new Dataface_Record('genesequences', array());
$record->setValues($defaultValues); #set the defaults for dataface
It looks like you are adding the same record to each entry in the array. This probably means that every row in your preview has the same values. -Steve
-
shannah
-
- Posts: 4457
- Joined: Wed Dec 31, 1969 5:00 pm
 by krazykit » Tue Dec 11, 2012 9:08 am by krazykit » Tue Dec 11, 2012 9:08 am
Thank you for your reply and pointing out the fix for the error in my code, but unfortunately, that was not my issue. Perhaps I was unclear: I am unable to set default values from the Xataface Import Records form itself: there are no drop-downs or text-fields to set defaults when importing from the web page. Please see the attached image.
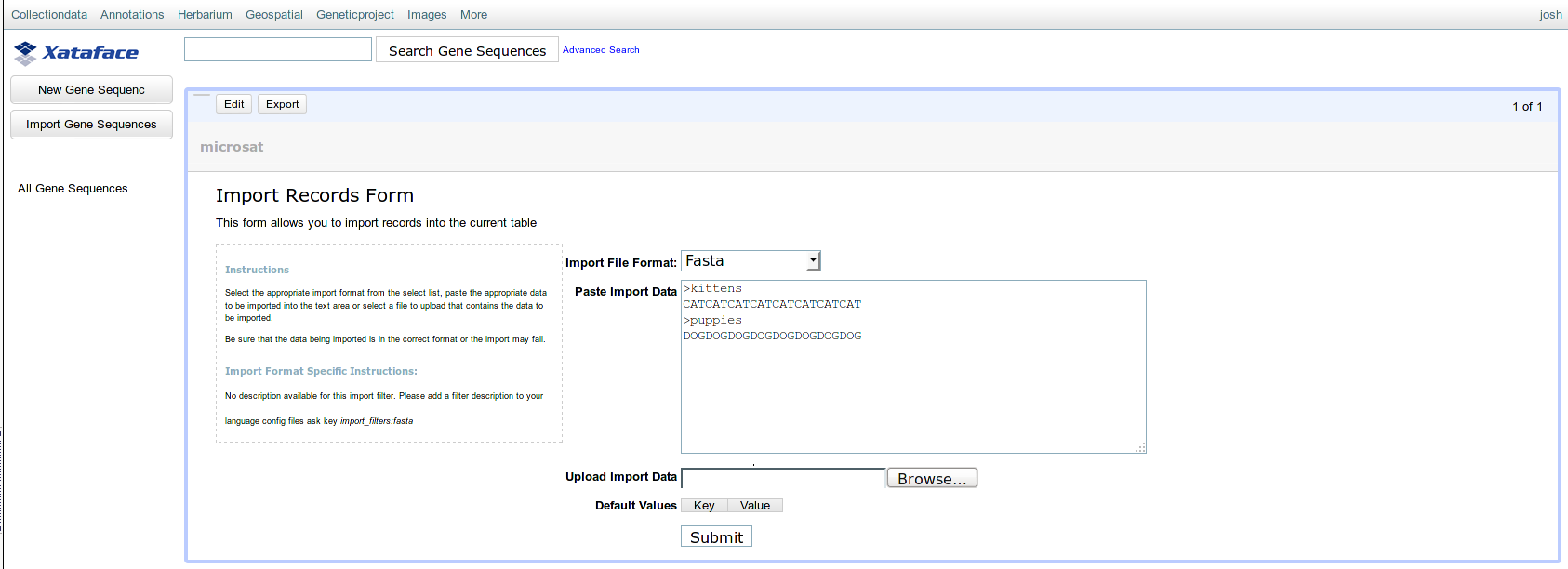 - import default value settings
- xataface.png (57.54 KiB) Viewed 9012 times
-
krazykit
-
- Posts: 4
- Joined: Mon Dec 10, 2012 10:28 am
 by shannah » Tue Dec 11, 2012 9:16 am by shannah » Tue Dec 11, 2012 9:16 am
Are there any javascript errors on that page? Are you using permissions/i.e. does this user have full permissions?
-Steve
-
shannah
-
- Posts: 4457
- Joined: Wed Dec 31, 1969 5:00 pm
 by krazykit » Tue Dec 11, 2012 9:53 am by krazykit » Tue Dec 11, 2012 9:53 am
Looks like the problem was on my end: apparently, my Adblock list was filtering out "addRow". Whitelisting the site (or removing the offending entry) fixes this. Importing now works as expected.
-
krazykit
-
- Posts: 4
- Joined: Mon Dec 10, 2012 10:28 am
Return to Xataface Users
Who is online
Users browsing this forum: No registered users and 15 guests

|
